During the harvest and ensiling processes, plant proteases are released, and they begin degrading the vegetative proteins even before they are fed. This is one reason ensiling practices emphasize the need to rapidly decrease silage pH to a point that it limits protease activity and stops protein degradation.
Variable and changing environmental conditions often lead to the production of less-than-optimal silage; protein degradation may be as high as 70 to 75 percent of the true protein.
And in the rumen of dairy cows, vegetative proteins of forages like alfalfa are rapidly degraded due to microbial proteases. Protein fragments (partially degraded protein molecules) are even less efficiently utilized, often being converted to urea.
Intact proteins are more effectively utilized, being converted to rumen microbial proteins of high nutritional value to the cow, but still, up to one-third of alfalfa protein is ultimately lost.
The impact of poorly utilized protein is both economic and environmental. Additional protein supplements that are much more expensive than forages must be purchased. And the unused protein nitrogen is excreted as urea; in the environment, this is readily lost to the atmosphere as ammonia.
Why red clover?
An exception to this often-observed problem with alfalfa and other forages, including corn silage, is red clover. Red clover, even under poor ensiling conditions, typically has minimal protein degradation, maintaining 70 to 80 percent of its protein intact. Thus, it provides more true protein in the diet and results in higher protein-use efficiencies.
This phenomenon was first recognized at the U.S. Dairy Forage Research Center (USDA Agricultural Research Service) in Madison, Wisconsin, when comparing alfalfa with red clover silage.
Upon further investigation, we determined that the key factor in red clover is an enzyme called polyphenol oxidase (PPO) and an abundance of its substrate, special chemicals called o-diphenols.
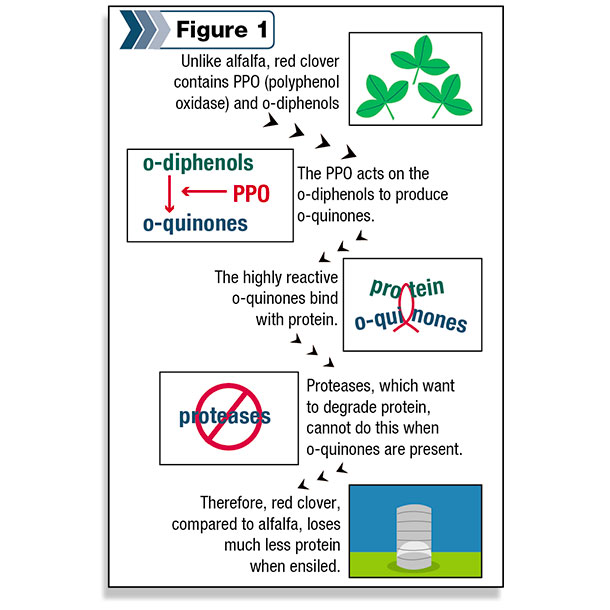
PPO converts the o-diphenols to o-quinones, which are highly reactive molecules that can bind to proteins (Figure 1).
These o-quinones react with the native proteases, decreasing their activity and leaving much of the protein intact during the ensiling process and in the rumen.
We then asked ourselves, “Can the PPO system that works so well in red clover be transferred to alfalfa?” The use of a PPO and o-diphenol system in alfalfa could enhance the utilization of alfalfa protein by reducing protein breakdown in the rumen, increasing plant protein flowing to the intestinal tract and increasing the absorption of amino acids in the intestinal tract.
To fully develop a PPO/o-diphenol system in alfalfa requires two new components to be introduced into alfalfa: an active PPO enzyme and the o-diphenols it needs as substrate to produce the critical o-quinones. An extensive survey of alfalfa germplasm indicated that there were no natural variants that contained active PPO in the vegetative portions of the plant.
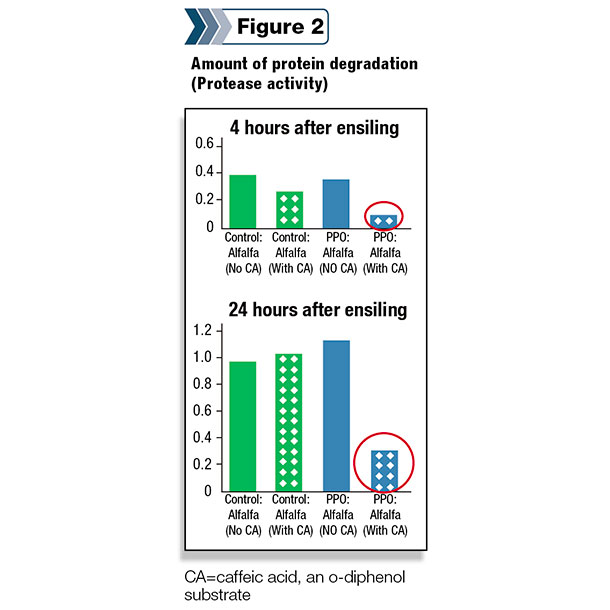
Therefore, a precision-breeding approach was used at the USDFRC to insert the red clover PPO gene into alfalfa. The first step of this process has been quite successful, and alfalfa plants expressing the red clover gene can inhibit proteolytic activity when appropriate o-diphenols are added as a substrate (Figure 2).
The next step of this process is to have a supply of o-diphenols, since alfalfa currently does not produce such PPO substrates. Of course, the most desirable approach would be to have alfalfa synthesize the appropriate o-diphenol substrate.
We are currently working on ways to introduce the necessary genes for the enzymes needed to produce a suitable o-diphenol in alfalfa. If this can be accomplished, the next step would be to develop high-yielding alfalfa cultivars that consistently produce desired types and quantities of o-quinones to protect the protein in alfalfa leaves.
An alternative approach would be to use external sources of diphenols that are abundant in many plants, such as potato peels, coffee grounds and forages like timothy and some varieties of tall fescue.
We are also working to test the feasibility of co-ensiling PPO forage with plant materials or plant extracts that contain o-diphenols to decrease proteolytic activity and preserve the greatest possible amount of the native protein as intact protein.
A feeding trial using cool-season grasses demonstrated that grasses with PPO alone (orchardgrass, smooth bromegrass) or with the o-diphenol substrate alone (tall fescue, timothy) did not increase the efficient use of the protein by lambs.
However, when the grasses were heavily conditioned and combined, native proteases were inhibited, more true protein was available to the lambs, and protein-use efficiency increased.
If an effective PPO/o-diphenol system is developed, we would then need to evaluate and optimize dairy cattle diets for this new alfalfa in order to enhance protein utilization and milk production.
We would also want to characterize nitrogen cycling as it goes from manure and terminated alfalfa stands to corn and other crops in order to optimize crop production practices and minimize nitrogen loss to the environment. FG
Michael Sullivan is a molecular geneticist with the U.S. Dairy Forage Research Center, USDA Agricultural Research Service.

-
Ronald Hatfield
- Plant Physiologist
- U.S. Dairy Forage Research Center
- USDA Agricultural Research Service
- Email Ronald Hatfield










